Bibliometric analysis of global trends in the study of the evolutionary history of chloroplasts in members of the Class Dinophyceae
Análisis bibliométrico de las tendencias globales en el estudio de la historia evolutiva de los cloroplastos en miembros de la clase Dinophyceae
Beatriz Lira1*
1Departamento de Ecología y Recursos Naturales, Facultad de Ciencias, Universidad Nacional Autónoma de México (UNAM). Ciudad Universitaria 04510, Ciudad de México. +52 55 5622 5430
Correspondencia: *bealirah@ciencias.unam.mx
Lira, B. 2021. Bibliometric analysis of global trends in the study of the evolutionary history of chloroplasts in members of the Class Dinophyceae. Cymbella 7(2): 51-64
Abstract
The analysis of the evolutionary relationships between members of the Class Dinophyceae and their chloroplasts hosts represents one of the keys to solving the complicated evolutionary history of chloroplasts acquisition in this group, and although the amount of work on the matter seems very scarce, in recent years it has been a significant development that could solve in the future evolutionary discrepancies in other groups. It is essential to analyze the trends within the study of this topic to understand the evolution of scientific literature as an essential step for developing and strengthening the field. The assessment was achieved by carrying out quantitative and qualitative analyses of global research and emerging trends from 1996 through 2020, using the Web of Science Core Collection. These analyses showed an increasing work on the study of the evolution of chloroplasts in dinoflagellates, and the leading countries were those with the most significant economic and scientific development. Likewise, even though many publications were analyzed in this work, the number of publications on this subject is still scarce compared to more general topics, so research will remain active and growing during the next decade.
Keywords: Bibliometric analysis, Dinophyceae, endosymbiosis, evolution.Resumen
El análisis de las relaciones evolutivas entre los miembros de la clase Dinophyceae y sus cloroplastos huéspedes representa una de las claves para resolver la complicada historia evolutiva de la adquisición de cloroplastos en este grupo, y a pesar de que la cantidad de información al respecto parece escasa, en los últimos años se ha realizado un avance importante que podría resolver en un futuro las discrepancias evolutivas en otros grupos. Para entender la evolución de la literatura científica como un paso fundamental para el desarrollo y fortalecimiento de este campo, es importante analizar las tendencias en el estudio del tema. Con este fin, se realizaron análisis cuantitativos y cualitativos de las investigaciones internacionales, así como de las tendencias emergentes durante el período 1996 - 2020, utilizando para ello la Web of Science Core Collection. Estos análisis mostraron una tendencia en el incremento de trabajos sobre la evolución de cloroplastos en dinoflagelados, con una clara tendencia al liderazgo de los países de mayor desarrollo económico y científico. Asimismo, a pesar de que en este trabajo se analizaron diversas publicaciones es evidente que el número de ellas sobre el tema es escaso, en comparación con otras más generales, por lo que se proyecta a futuro que la investigación sobre los cloroplastos en el grupo estará activa y en crecimiento durante la próxima década.
Palabras clave: Análisis bibliométrico, Dinophyceae, endosimbiosis, evolución.
Introduction
Endosymbiosis has been the most important event in the history of the emergence and evolution of photosynthesis, without which life, as we know it today, would not exist. Over the eons, endosymbiosis of a cyanobacterium by a eukaryotic organism that resulted in the origin of the primary plastids and their subsequent lateral transfer to various eukaryotic lineages, giving rise to secondary plastids, has drawn the attention of many researchers (Bhattacharya & Medlin 1998; Cavalier-Smith 1999; Delwiche 1999; Gabrielsen et al. 2011).
However, complex plastids originated from endosymbiosis between eukaryotes that have incorporated secondary plastids and gave rise to tertiary plastids, which have not been well studied (Inagaki et al. 2000). These tertiary plastids are characterized by presenting three to four membranes and modifications of intracellular transport (Archivald 2009; Gray & Spencer 1996). In this context, dinophytes are a unique group that can present both secondary and tertiary chloroplasts that have been acquired by various algal lineages (Gabrielsen et al. 2011), incorporating organisms such as diatoms which already have chloroplasts of secondary origin (Calassan et al. 2018; Hehenberger et al. 2014; Keeling 2004, 2010). These dinophytes have been commonly called dinotoms (Imanian et al. 2010).
Based on the ultrastructural analysis of dinotoms, it has been observed that the endosymbiont retains its photosynthetic machinery, as well as a functional nucleus with protein-coding genes, a considerable amount of cytosol, and mitochondria (Imanian & Keeling 2007; Imanian et al. 2012; McEwan & Keeling 2004; Takano et al. 2008). Despite the degree of independence and minimal reduction, the endosymbiont diatom is present in all stages of development of the host cell, indicating a synchronization between their life cycles (Calassan et al. 2018), unlike the ancestral peridinin chloroplasts of the dinophytes. These chloroplast remnants have been considered a unique endosymbiont stigma (Dodge 1983; Figueroa et al. 2009; Horiguchi et al. 1999; Moestrup & Daugbjerg 2007; Takano et al. 2008), surrounded by a triple membrane structure (Horiguchi & Pienaar 1994).
Molecular analysis of the transcriptome of the Durinskia baltica Carty & E.R. Cox 1986, and Kryptoperidinium foliaceum (F. Stein) Lindemann 1924 have revealed that almost no functional reduction has occurred in the nucleus of their diatoms. However, these studies have demonstrated the independence between the diatom and the host; the latter has managed to maintain the endosymbiont by controlling its karyokinesis (Hehenberger et al. 2016). Based on these studies, it has been suggested that diatoms of dinotoms constitute an intermediate phase of evolution between kleptoplasty and incorporated chloroplasts (Horiguchi 2006; Keeling 2010; Yamada et al. 2019).
Although analyses of the evolutionary relationships between dinotoms represent one of the keys to solving the tangled evolutionary history between hosts and endosymbionts in this group (Matsuo & Inagaki 2018), the number of studies on this subject seems scarce, so it is essential to analyze the tendency of these studies to understand the evolution of the scientific literature as an essential step for the development and strengthening of this study field.
A useful tool for this type of analysis is bibliometrics. This is a research method used within the study of information sciences which includes quantitative and statistical analyses (Sun et al. 2011). These analyses have become fundamental tools for studying current trends within the scientific literature and in specific areas (Oliveira et al. 2020).
A common approach to bibliometrics is the Science Citation Index (SCI), which tracks publications from the Institute for Scientific Information (ISI). The bibliometric analyzes carried out using the ISI have been widely used in topics related to photosynthesis (Yu et al. 2012), research on microalgae (Rumin et al. 2020), diatoms (Zhang et al. 2019), and dinoflagellates (Oliveira et al. 2020). And recently, the use of different data bases such as Coci, Scopus, Lens and Semantic scholar is also recommended. However, although these studies indirectly address the evolution of dinotoms, this area has not been emphasized. Due to this situation, the main objective of this work is to analyze global research and emerging trends quantitatively and qualitatively in studies about the evolution of dinotoms to provide objective guidelines and trends for future research.
Methods
Data were obtained from the Thomson Reuters Web of Science™ Core Collection database, including the Science Citation Index Expanded bases (SCIE): SSCI, A & HCI, CPCI-S, CPCI-SSH, BKCI-S, BKCI-SSH and ESCI.
A detailed search was carried out proving different sets of the keywords and the final set was decided upon the one that produced the most entries falling within the main subject related to the evolution of chloroplast in dinophytes. The final set was obtained using the following keywords and Boolean operators: “dinoflagellate OR dinophyte” AND “endosymbiont” AND “plastid” OR “chloroplast”, the word “evolution” was not including in this set because it produced entries related to more general subjects. All documents from 1996-2020 were included. One hundred sixty-nine entries coincided with the established criteria, of which 25 were removed because they corresponded to groups other than dinoflagellates. The final database was of 144 records. Obtained information included data on authors, title, origin, abstract, year of publication, countries, type of document, keywords, citations, and subject category. Data was exported in simple text format of the ANSI type (FN ISI Export Format VR).
The publications obtained from the databases were organized and processed using the Bibliometrix 3.0 package (Aria & Cuccurullo 2017) from RStudio. Data processing, statistical analysis, and graphics generation were carried out with BiblioShiny application of the Bibliometrix 3.0 package. The bibliometric analysis was divided into three parts: an intellectual structure, a conceptual structure, and a social structure analysis of the information.
To understand the intellectual structure of the information, a co-citation analysis of the information was carried out, reviewing the scientific production over time, as well as the total number of citations per year. The relationship between the countries with the most significant scientific production was analyzed as well as the thematic evolution.
The conceptual structure was evaluated with a word grouping analysis in which the evolution of trending topics and keywords dynamics were reviewed. Bradford’s model was used to estimate the exponential decrease in performance when expanding the search for references in scientific journals, as well as the dynamics of the references.
The evaluation of the social structure was performed with a co-authorship analysis, within which the impact of the 50 most cited authors was determined based on the number of citations their articles received (h-index), the most relevant affiliations, and global distribution. According to their productivity, the Lotka model was determined as well as a co-authorship network.
Results
The 144 publications resulting from the initial search in the Thomson Reuters Web of Science ™ database comprised 122 articles (85 %), two book chapters (1 %), six proceedings articles (4 %), one editorial material (1 %), ten reviews (7 %), and three book article reviews (2 %). All 144 publications were in English (100 %).
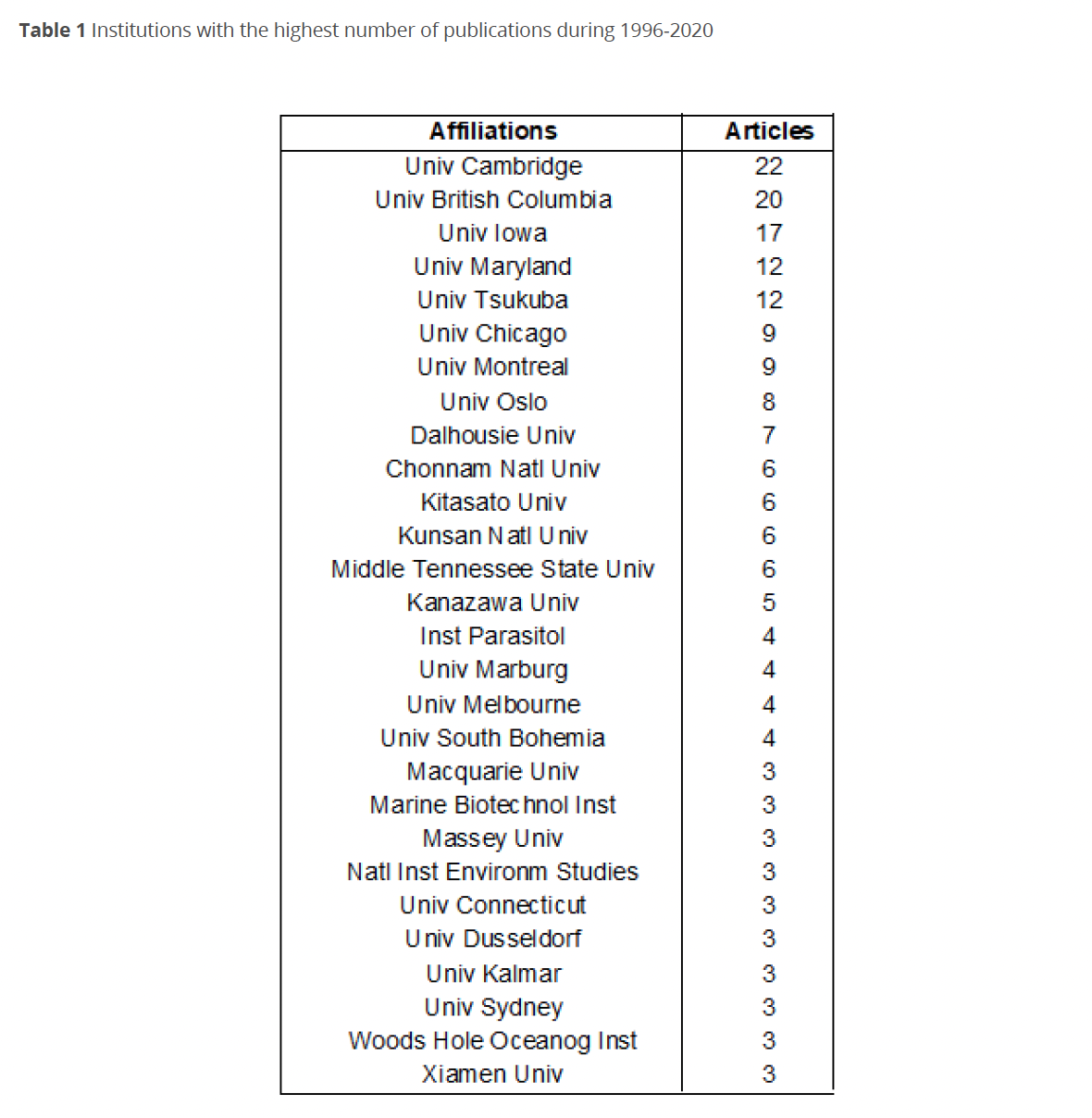
Figure 1 shows the trend in the number of publications changes during 1996-2020. From 1996-1998, the production was poor, while for 1999 there was a considerable increase to six publications. Subsequently, a gradual increase in publications was observed with three in 2000, five in 2001 and seven in 2002, decreasing again to four in 2003.
In the following four years, we could see progress, with the largest number of publications: nine in 2004 and 2006, 10 in 2007, and eight in 2008, with 2005 being the best year for the evolution of dinotoms research, with 11 publications. From 2009 to 2014, there was a fluctuation, and 2011 was the year with most publications in the period.
From 2015 to 2017, production remained low, and in 2018 it increased to seven publications. However, during 2019 and 2020, production dropped. An exponential regression model was adjusted to the data of the analyzed period (R2 = 0.705), showing that the trend in the number of publications will continue with an average of six publications for the next ten years.
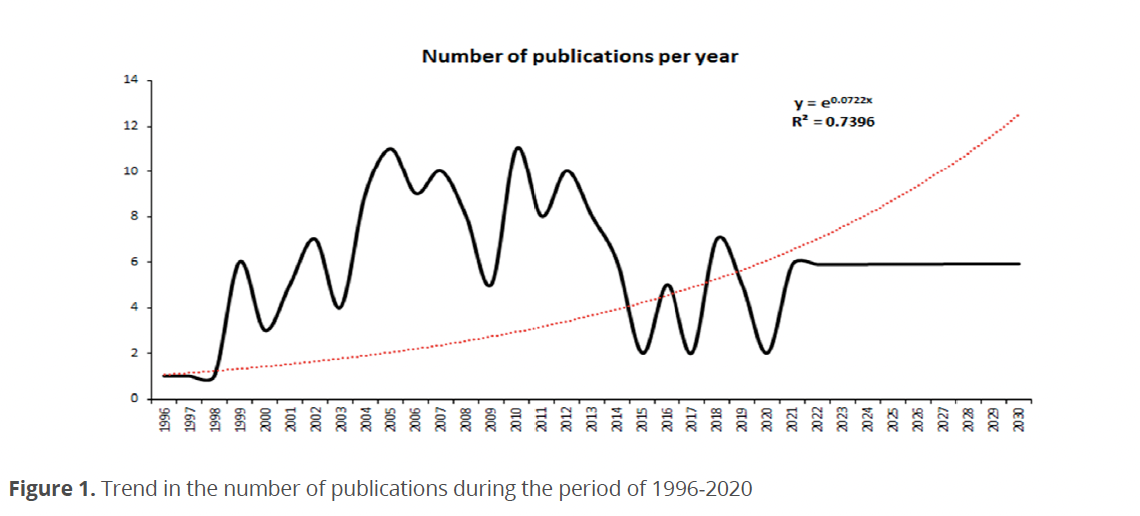
Regarding the relationship in the number of citations per year (Fig. 2), a stable trend was noted between 1996-1998, with one citation in 1999 followed by an increase of six citations. In the following four years, a fluctuation between three and seven citations was observed.
From 2004-2007 a positive trend was observed with nine to 11 citations. The last value corresponded to 2005, the year with the highest number of citations in the interval. There was a fluctuation from 2008-2013, with ten citations in 2010-2012 and eight in 2011-2013. In 2014 and 2015, there was a decrease with six and two citations, respectively. From 2016 to 2020, there were no more records.
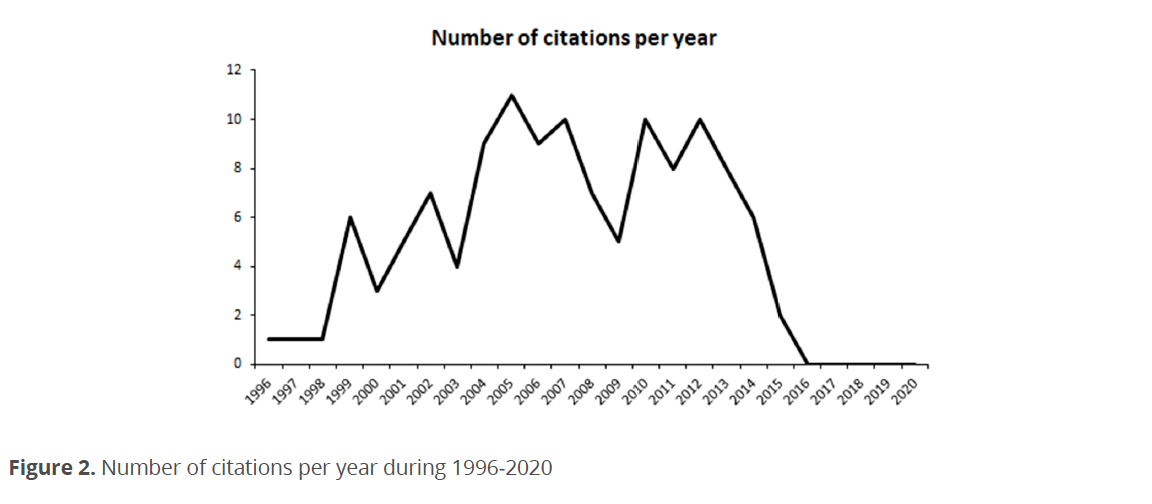
Figure 3 shows how the relationships between the thematic of chloroplasts evolution research in dinophytes have changed over time. In the first cut that comprised the 1996-2008 period, it was observed that the topics related to maximum likelihood analysis for the study of chloroplast genomes had the least number of publications in that period, and in the second cut, corresponding to the period 2009-2020, there was no increase in publications or thematic relationships.
The next topic with the least number of publications is related to the study of ribosomal genes, which for the second section was related to topics of diversity and chloroplast genomes. The topic “chloroplast” was related to the topic “apicomplexan parasites” towards the second cut, as well as with dinoflagellates in general.
The evolution topic presented a relationship with a greater number of publications about chloroplast genomes. Finally, the topic with the highest number of publications was “dinoflagellates” and relating to this topic of the second cut with sequencing and diversity.
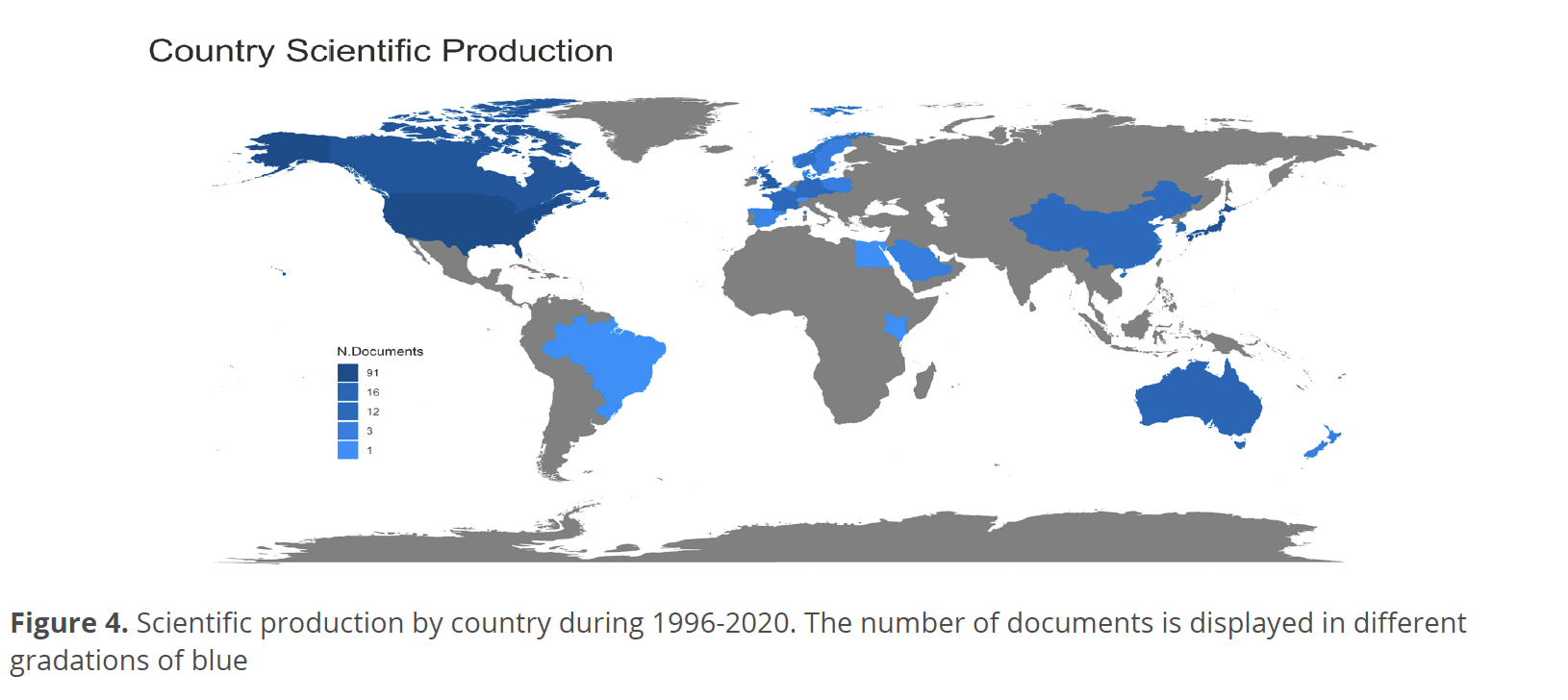
Figure 4 shows the relationship of the largest scientific production by country during 1996-2020. It was observed that the largest production is led by the USA, United Kingdom, and Japan, followed by Canada, China, France, Germany, Norway, and Australia. In third place are Sweden, Poland, Belgium, Spain, and Saudi Arabia. In the last places are Egypt, Kenya, and Brazil.
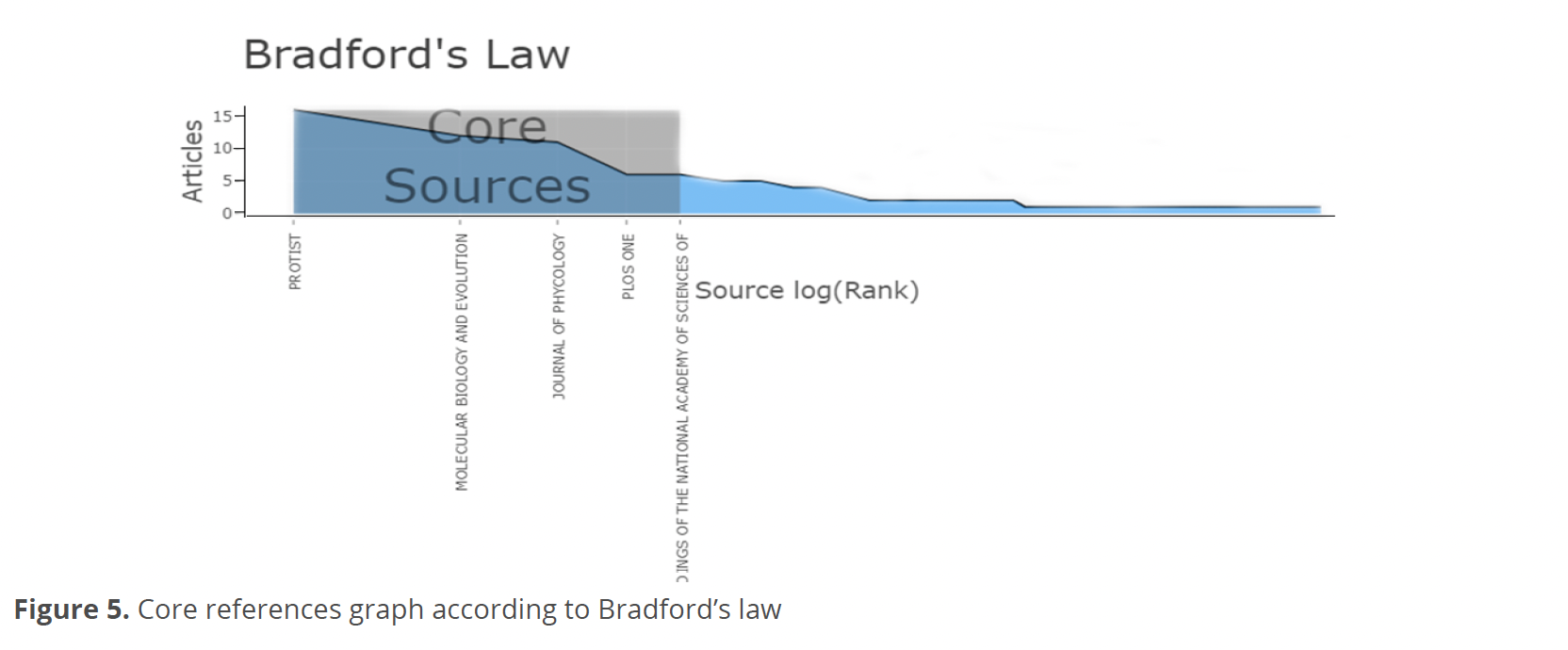
About the conceptual structure, based on Bradford’s law, the journals Protist, Molecular Biology and Evolution, Journal of Phycology, PloS One and Proceedings of the National Academy of Sciences of the United States of America were identified as the core references (Fig. 5).
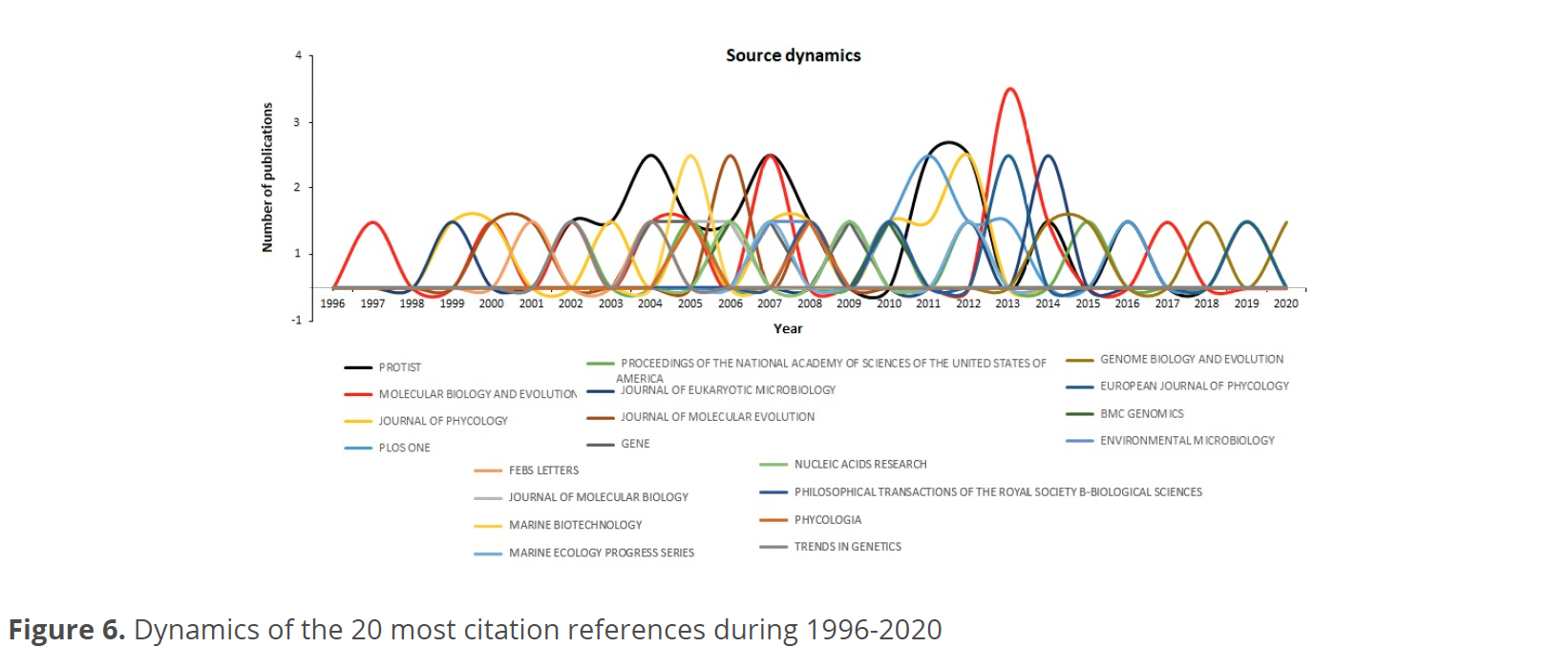
Figure 6 shows the dynamics of the 20 most cited references during 1996-2020. Most of them followed the same trend over time, oscillating between 0-1 publications. The sources that stand out with two publications are Marine Biotechnology in 2005, Journal of Molecular Evolution in 2006, PloS One in 2011, Journal of Phycology in 2012, European Journal of Phycology in 2013, and the Journal of Eukaryotic Microbiology in 2014. The journal Molecular Biology and Evolution issued two publications in 2007 and three in 2013; however, Protist leads the sources with two publications in 2004, 2007, 2011, and 2012.
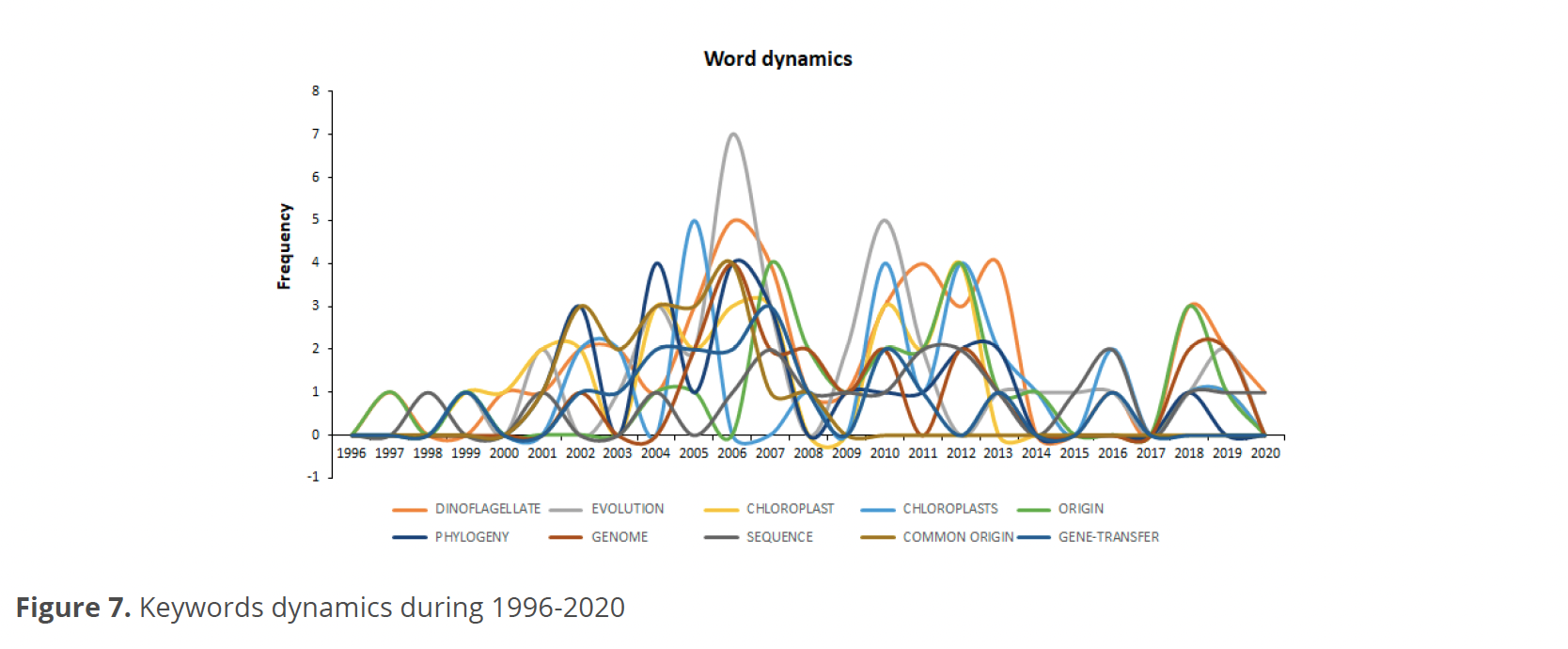
Concerning keywords dynamics over 1996-2020, Fig. 7 shows that the words with the highest frequency were “evolution”, with its maximum values in 2006-2010, and “dinoflagellates” in 2006, 2011, 2013, and 2018. The terms “chloroplast” and “phylogeny” presented the greatest fluctuations in frequency over time, the first with the highest frequency records in 2005, 2010, and 2012. For its part, the term “phylogeny” presented the highest frequency values in 2004 and 2006. The word “origin” presented the highest frequency during 2007 and 2012.
Figure 8 shows four main clusters: the first one, in red, corresponded to “evolution” which was the central word closely related to “sequence”, “chloroplast genomes”, “ribosomal RNA” and “DNA”. The words “RNA”, “nucleus”, “plastid genome”, “expression”, “organization”,” Amphidinium operculatum” “dinoflagellate”, “non-coding region”, “RNA gene transfer” and “Minicircles” had a weaker relation with the central word “evolution”.
The second cluster in purple, corresponded to the word “dinoflagellate” as the central word with a close relationship with “chloroplast”, “phylogeny”, “genome”, and “origin”. These words in turn are related in a lesser extent with “plastids”, “apicomplexan”, “genes”, “diversity”, “kleptoplasty”, “molecular evidence”, “acuminata”, “Myrionecta rubra” and “retention”.
The third cluster in blue, showed “common origin” as the central word closely related to “protein” and “gene transfer”. The words “single origin”, “maximum likelihood”, “red algae”, “dinoflagellate plastids”, “endoplasmic reticulum” and “Plasmodium falciparum” were observed with a lesser relationship. The last cluster in green, presented weak relationships between the words “ultrastructure”, “green dinoflagellate”, “fine structure”, “endosymbiont” and “marine dinoflagellate”.
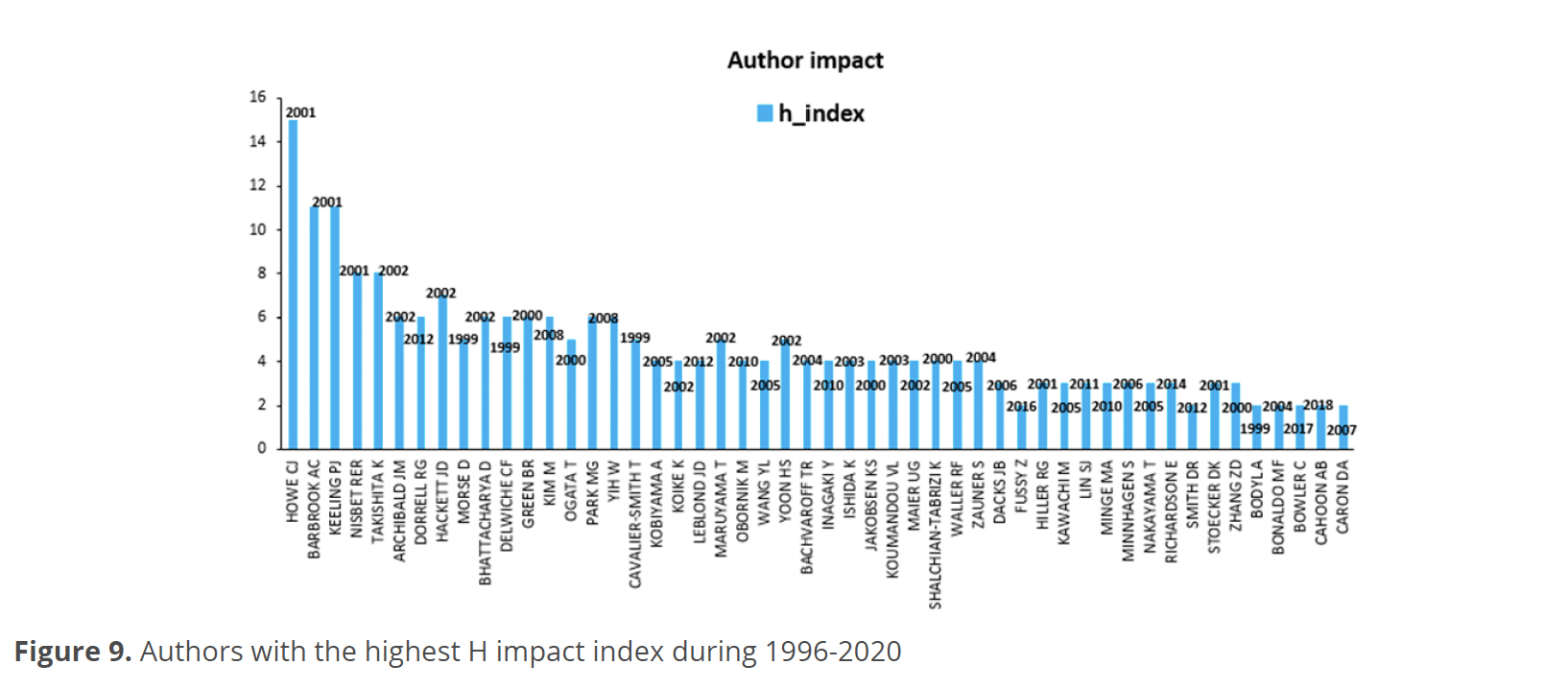
For the social structure, Fig. 9, shows the 50 most cited authors based on their h impact indexes, Howe presented an H index of 15 followed by Barbrook and Keeling, both with an index of 11. Nisbet and Takishima presented an index of eight, Hackett seven, Archibald, Dorrell, Bhattacharya, Delwiche, Green, Kim, Park, Yih with an index of six. For their part, Morse, Ogata, Cavalier-Smith, Maruyama, and Yoon presented an index of five. Kobiyama, Koike, Leblond, Obornik, Wang, Bachvaroff, Inagaki, Ishisa, Jakobsen, Koumandou, Maier, Shalchian-Tabrizi, Waller and Zauner had an index of four. With an index of three Dacks, Hiller, Kawachi, Lin, Minge, Minnhagen, Nakayama, Richardson, Stoecker, and Zhang. Finally, Fussy, Smith, Bodyl, Bonaldo, Bowler, Cahoon and Caron are grouped with an index of two.
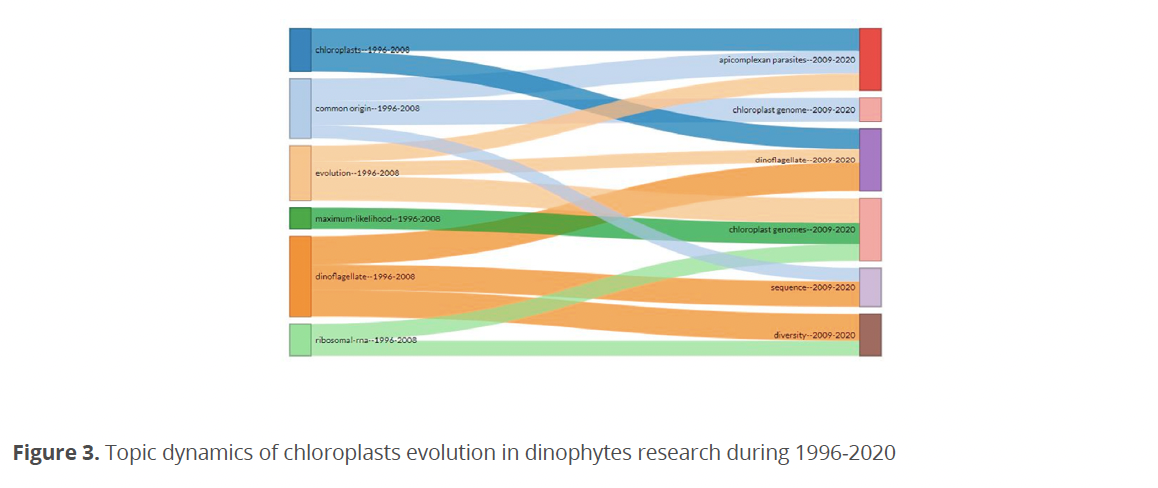
Table 1 shows the most relevant institutions and their number of publications during 1999-2020. In this analysis, the University of Cambridge stands out with 22 publications followed by the Universities of Iowa with 17, Maryland and Tsukuba both with 12 publications respectively. The Universities of Chicago and Montreal presented nine publications, while the Universities of Oslo eight and Dalhousie seven.
Chonnam Natl, Kitasato, Kunsan Natl, and Middle Tennessee State Universities submitted six publications. Kanazawa University had five publications. The Parasitol Institute and the Universities of Marburg, Melbourne, South Bohemia presented four. With three publications, the Universities of Macquarie, Massey, Connecticut, Dusseldorf, Kalmar, Sidney, and Xiamen stood out, as well as the Marine Biotechnology Institutes, Natural Environmental Studies, and the Oceanographic Woods Hole.
A direct relationship was observed between the countries with the highest production and the most relevant institutions. From the 136 institutions analyzed (Table 1) only five presented more than ten publications during the analyzed period, grouping 27 % of the total production. The institution with the highest production is in England (Cambridge Univ.), followed by a Canadian institution (British Columbia Univ.). Although the number of publications from US institutions is lower, this country has a higher representation because it has more institutions within the list (Univ. Iowa and Univ. Maryland). Finally, the last place within the five most productive institutions is Japan (Univ. Tsukuba).
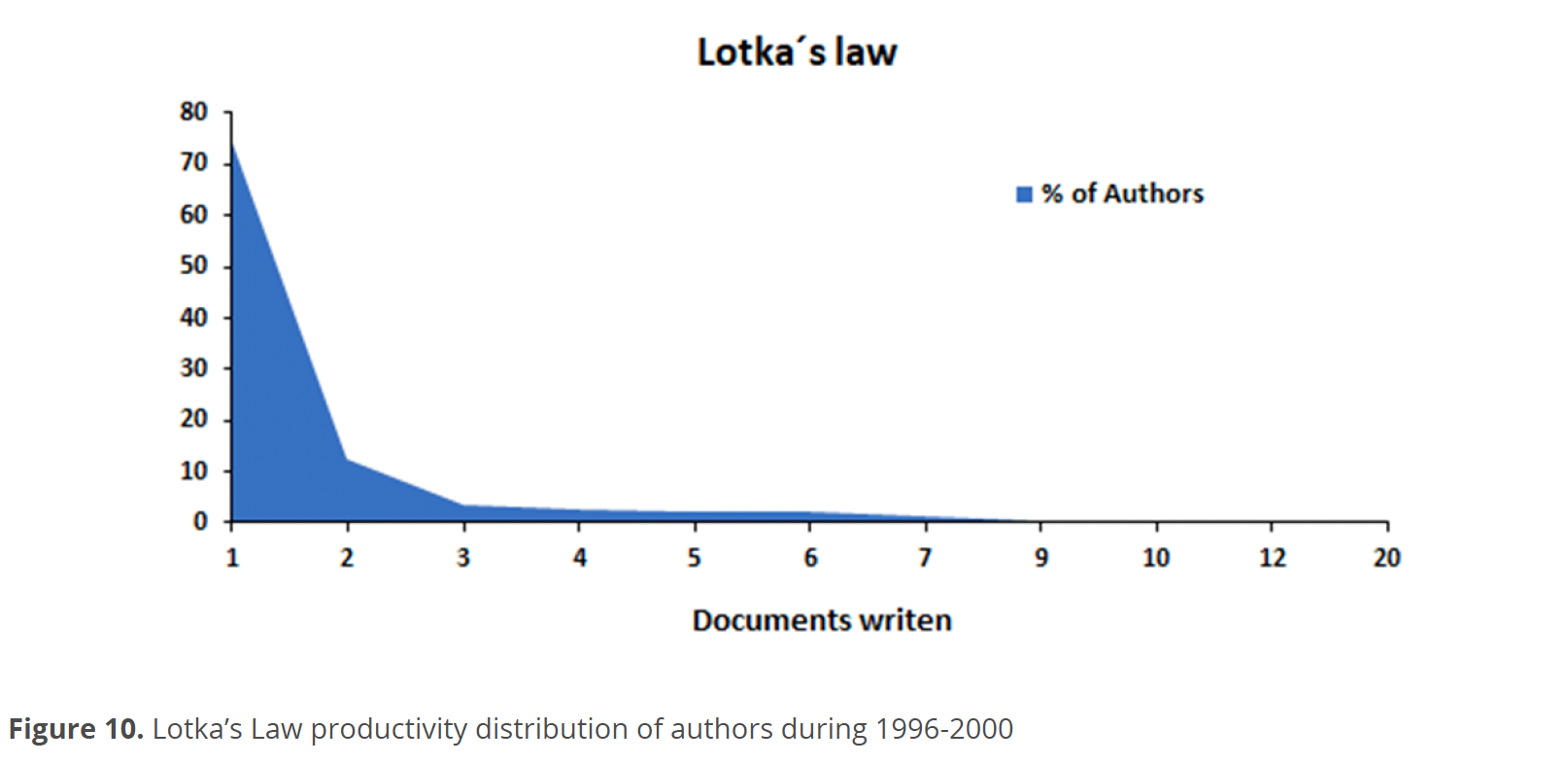
Based on the results of Lotka’s Law during 1999-2020, Fig. 10 shows that in the distribution of authors according to their productivity, 74 % had 1 publication, 12 % had 2 publications, 3 % had 4, 2.4 % had 3, 2 % between 5-6 and only 1 % had between 7-12 publications.
In Fig, 11, ten collaboration clusters were observed, among which Howe’s cluster (1) stands out with eight collaborations, the largest collaborations were between Howe – Nisbet, Howe – Barbrook, and Howe – Dorell. The weaker collaborations in this cluster were the ones with Howe – Koumandou, Richardson, Dacks, Bowler, and Hiller. The next major cluster was the Takishita (2), which features the largest collaboration with Kobiyama, Koike, Ogata, and Maruyama. On the other hand, the weaker collaborations were between Takishita and Kawachi, Inagaki and Nakayama. In Keeling cluster (3), a closer collaboration with Archibald and more distant with Waller, Obornil, and Fussy was observed.
Cluster 4 presented a close collaboration between Bhattacharya and Hackett and less with Yoon and Bonaido. In cluster 5, a network was observed between Park, Yih, and Kim. Jakobsen’s cluster (6), showed a close collaboration between Shaichian -Tabrizi and Minge. In Green’s cluster (7), a close collaboration with Cavalier-Smith and Zhang was observed. On the other hand, small collaboration networks between two authors were observed, as in the case of Wang-Morse and Bachvaroff-Delwiche. The analysis did not group the authors Lin, Leblond, Cahoon, Smith, Ishida, Stoecker, Minnhagen, Bodyl, and Caron.
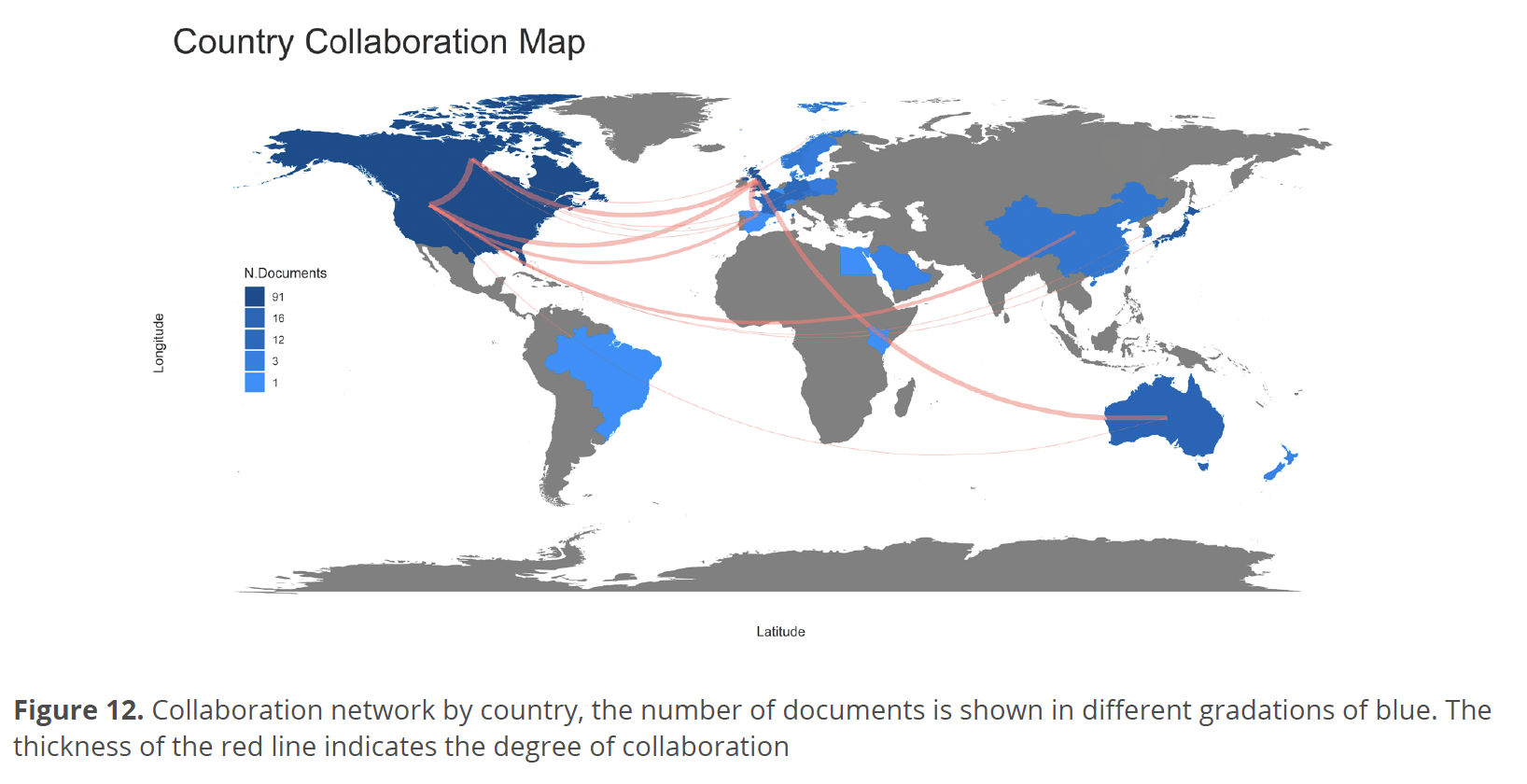
Figure 12 shows the map of the collaborations network by country where the country that stood out with the strongest collaborations was the USA connected with Canada, China, United Kingdom, and Spain, with a weaker collaboration USA connected with Australia and Japan. On the other hand, it was observed that Canada presented collaborations with Norway, Germany, the Czech Republic, and Spain.
Discussion
Within the intellectual structure (Figs. 1-3) during the period of lower production, most of the publications focused on the study of the diversity of chloroplasts in dinoflagellates, their possible origins, and different evolutionary lines related to organisms with similar characteristics for classification purposes (Baldauf 2003; Dewilche 1999; Saldarriaga et al. 2001; Zhang et al. 1999). This period was followed by the years of the highest publication between 2004-2007, during which new tools and techniques were developed, especially molecular ones that allowed a greater approach to phylogeny issues not only for classification purposes but also with a diversification towards physiological and evolutionary topics within which they begin to approach to endosymbionts as an important part of the origin of chloroplasts (Horiguchi 2006; Koike et al. 2005; Moestrup & Daugbjerg 2007; Okamoto & Inouye 2006; Pearce & Hallegraeff 2004).
This development is observed in the thematic evolution of phylogenetic analyses with the incorporation of maximum likelihood analysis and work with ribosomal RNA (Fig. 3). In this context, the increase in production during 2010-2014 may be related to the development of molecular techniques that allowed progress towards the study of the origin of chloroplast diversity in dinoflagellates focused on endosymbionts (Imanian et al. 2010; Keeling 2010; Stoecker et al. 2009; Takano et al. 2014). For the last years of the 2010-2020 decade, the study of the relationships between the origin of chloroplasts and the evolutionary history of endosymbionts has been refined (Bengtson et al. 2017; Čalasan et al. 2017; Cavalcante et al. 2017; Gottsching et al. 2017; Hehenberger et al. 2016; Janouškovec et al. 2017; Yamada et al. 2017).
Even though in 2019 was an increase in production (Lira & Tavera 2019; Yamada et al. 2019) and the trend seemed to be positive, production fell sharply in 2020. This trend was repeated in the number of citations per year related to the number of publications per year (Fig. 2).
Although there is still no statistical information on the reason for this drop, it is likely due to the difficulties that the Covid-19 pandemic has imposed on scientific research worldwide. These difficulties range from the impossibility of entering work centers to field and laboratory work. The statistical relationship of annual publication production showed a positive trend with a spike in publications for the next ten years. However, this trend may be underestimating the effects, at different scales, derived from the pandemic on research on this topic due to the interruption of international collaboration research.
The world distribution of scientific production was concentrated among the USA, England, and Japan (Fig. 4), focusing on particular working groups. This distribution can be related to social and environmental aspects, but above all, economic, and in this context, the USA has been for many years the most productive country with the most significant influence in different areas of research (Oliveira et al. 2020).
Despite this, the dynamics of production distribution have been changing, incorporating communities from other countries in Europe, Asia, and Africa, mainly due to the development of collaboration networks among researchers from countries with the highest production and working groups in countries with lower production (Fig. 12).
The conceptual structure of this analysis showed that most of the information analyzed is concentrated in 5 journals (Fig. 5). These journals do not coincide entirely with those leading the reference dynamics (Fig. 6). The reason for this difference is that there are journals within the core, such as Protist that cover a wide diversity of topics. For this reason, this journal presents few publications for more years, unlike journals such as Molecular Biology and Evolution that have more publications but only for a specific couple of years. In this context, it is interesting to note that the journal PloS One, although it is among the most cited, being open access and covering basic research topics in any matter related to science and medicine, only had 2 publications on this topic in a year (2011).
The word dynamics match the thematic evolution of the terms “evolution” and “dinoflagellate”, which are central terms in all word analyses. These terms formed thematic networks that connect other words like “chloroplast” and “phylogeny”, being these terms the ones that had more significant fluctuations in frequency over time, increased during the years of greatest publication.
Despite the words “chloroplast” and “phylogeny” continue to appear within a taxonomic and classification context as suggested by Oliveira et al. (2020), they have been increasing their relevance within the networks formed by the central words, indicating that these concepts have acquired relevance in the subject of evolution.
The analyses of the social structure showed that the increases in the number of publications in European countries are mainly due to the network of collaborations with scientists from institutions and countries with the highest production; the most extensive collaborations have been carried out among scientists with a high level of production impact index (Figs. 9-11) although these have formed close regional associations (Fig. 12). Scientists in Asia have formed almost no international collaborations and concentrated in small local groups that have grown in recent years. This study does not reflect the dynamics of collaboration with countries that publish in regional or local journals, even though they work with scientists with a high level of impact. On the other hand, it will be important to analyze the changes in collaboration networks and their effects on international production in the context of the pandemic.
This bibliometric analysis showed a trend towards the increase in publications about the evolution of chloroplasts in dinoflagellates from 1996 to 2020. The countries with the highest production are countries with the greatest economic and scientific development, among which the United States of America and England are the most intertwined within the global research network together with Japan.
Even though many publications were analyzed in this work, the number of publications on this subject is scarce compared to more general areas, so research on this topic will remain active and growing in the future. A clear trend was observed towards research on the evolution of chloroplasts in dinoflagellates, based on the use of molecular tools increasingly accessible to most communities, together with the study of phylogenetic controversies related to gaps in taxonomic information and classification. The lack of information on the evolution of chloroplasts in Dinophyceae will be solved by fostering international cooperation among the most productive and resourceful countries and researchers with working groups in less developed countries.
References
Archivald, J.M. 2009. The Puzzle of Plastid Evolution. Current Biology 19: 81-88.
Aria, M. & C. Cuccurullo. 2017. Bibliometrix: An R-tool for comprehensive science mapping analysis. Journal of Informetrics 11: 959-975.
Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300: 1703-1706.
Bengtson, S., T. Sallstedt, V. Belivanova & M. Whitehouse. 2017. Three-dimensional preservation of cellular and subcellular structures suggests 1.6-billion-year-old crown-group red algae., PLOS Biology https://doi.org/10.1371/journal.pbio.2000735.
Bhattacharya, D. & L. Medlin. 1998. Algal phylogeny and the origin of land plants. Plant Physiology 116: 9-15.
Čalasan, A.Ž., J. Kretschmann & M. Gottschling. 2017. Absence of co-phylogeny indicates repeated diatom capture in dinophytes hosting a tertiary endosymbiont. Organisms Diversity & Evolution 18: 29-38.
Cavalcante, K.P., S.C. Craveiro, A.J. Calado, T.A.V. Ludwig & L. de S. Cardoso. 2017. Diversity of freshwater dinoflagellates in the State of Paraná, southern Brazil, with taxonomic and distributional notes. Fottea 17: 240-263.
Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellates, and sporozoan plastid origins and the eukaryotic family tree. Journal of Eukaryotic Microbiology 46: 347-366.
Delwiche, C. F. 1999. Tracing the thread of plastid diversity through the tapestry of life. The American Naturalist 154: 164-177.
Dodge, J. D. 1983. The functional and phylogenetic significance of dinoflagellate eyespots. Biosystems 16: 259-267.
Figueroa, R. I., I. Bravo, S. Fraga, E. Garces & G. Llaveria, 2009. The life history and cell cycle of Kryptoperidinium foliaceum, a dinoflagellate with two eukaryotic nuclei. Protist 160: 285-300.
Gabrielsen, T.M., M.A. Minge, M. Espelund, A. Tooming-Klunderud, V. Patil, A.J. Nederbragt, C. Otis, M. Turmel, K. Shalchian-Tabrizi, C. Lemieux, & K.S. Jakobsen. 2011. Genome Evolution of a Tertiary Dinoflagellate Plastid. PLoS ONE 6: e19132. doi:10.1371.
Gottsching, M., A. Ž. Čalasan & J. Kretschmann. 2017. Two new generic names for dinophytes harbouring a diatom as an endosymbiont, Blixaea and Unruhdinium (Kryptoperidiniaceae, Peridiniales). Phytotaxa 306: 296-300.
Gray, M.W. & D.F. Spencer. 1996. Organellar evolution. In: D.M., Roberts, P. Sharp, G. Alderson, & M.A. Collins. Eds. Evolution of Microbial Life. Cambridge University Press, Cambridge.
Hehenberger, E., B. Imanian, F. Burki & P. J. Keeling. 2014. Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts. Genome Biology and Evolution 6: 2321-2334.
Hehenberger, E., F. Burki, M. Kolisko & P. J. Keeling. 2016. Functional relationship between a dinoflagellate host and its diatom endosymbiont. Molecular Biology and Evolution 33: 2376-2390.
Horiguchi, T. & R.N. Pienaar. 1994. Ultrastructure and ontogeny of a new type of eyespot in dinoflagellates. Protoplasma 179: 142-150.
Horiguchi, T. 2006. Algae and their chloroplasts with particular reference to the dinoflagellates. Paleontological Research 10: 299-309.
Horiguchi, T., H. Kawai, M. Kubota, T. Takahashi & M. Watanabe. 1999. Phototactic responses of four marine dinoflagellates with different types of eyespot and chloroplast. Phycological Research 47:101-107.
Imanian, B. & P.J. Keeling. 2007. The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evolutionary Biology 7: 172.
Imanian, B., J. F. Pombert & P. J. Keeling. 2010. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One 5: e10711.
Imanian, B., J. F. Pombert, R.G. Dorrell, F. Burki & P. J. Keeling. 2012. Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One 7: e43763.
Inagaki, Y., J.B. Dacks, W.F. Doolittle, K.I Watanabe & T. Ohama. 2000. Evolutionary relationship between dinoflagellates bearing obligate diatom endosymbionts: insight into tertiary endosymbiosis. International Journal of Systematic and Evolutionary Microbiology 50: 2075-2081.
Janouškovec, J., G.S. Gavelis, F. Burki, D. Dinh, T.R. Bachvaroff, S.G. Gornik, K.J. Bright, B. Imanian, S.L. Strom, F. Delwiche, R.F. Waller, R.A. Fensome, B.S. Leander, F.L. Rohwer & J.F. Saldarriaga. 2017. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proceedings of the National Academy of Sciences 114: 171-180.
Keeling, P. J. 2004. Diversity and evolutionary history of plastids and their hosts. American Journal of Botany 91, 1481-1493.
Keeling, P. J. 2010. The endosymbiotic origin, diversification, and fate of plastids. Philosophical Transactions of the Royal Society 365: 729-748.
Koike, K., H. Sekiguchi, A. Kobiyama, K. Takishita, M. Kawachi, K. Koike & T. Ogata. 2005. A novel type of kleptoplastidy in Dinophysis (Dinophyceae): presence of haptophyte-type plastid in Dinophysis mitra. Protist 156: 225-237.
Lira, B. & R. Tavera. 2019. Life history and cell cycle of Durinskia baltica (Dinophyceae: Peridiniales) in culture. Nova Hedwigia 108: 37-50.
Matsuo, E. & Y. Inagaki. 2018. Patterns in evolutionary origins of heme, chlorophyll a and isopentenyl diphosphate biosynthetic pathways suggest nonphotosynthetic periods prior to plastid replacements in dinoflagellates. PeerJ 6: e5345.
McEwan, M.L. & P.J. Keeling. 2004. HSP90, tubulin and actin are retained in the tertiary endosymbiont genome of Kryptoperidinium foliaceum. Journal of Eukaryotic Microbiology 51: 651-659.
Moestrup, Ø. & N. Daugbjerg. 2007. On dinoflagellate phylogeny and classification. In: J., Brodie & J. Lewis. Eds. Unravelling the algae. The past, present and future of algal systematics. CRC Press, Boca Raton.
Okamoto, N. & I. Inouye. 2006. Hatena arenicola gen. et sp. nov., a katablepharid undergoing probable plastid acquisition. Protist 157: 401-419.
Oliveira, C.Y.B., C.D.L. Oliveira, M.N. Müller, E.P. Santos, D.M.M. Dantas & A.O. Gálvez. 2020. A scientometric overview of global dinoflagellate research. Publications 8: 50.
Pearce, I. & G.M. Hallegraeff. 2004. Genetic affinities, ecophysiology and toxicity of Prorocentrum playfairii and P. foveolata (Dinophyceae) from Tasmanian freshwaters. Phycologia 43: 271-281.
Rumin, J., E. Nicolau, R. Gonçalves de Oliveira Junior, C. Fuentes-Grünewald, K.J. Flynn & L. Picot. 2020. A bibliometric analysis of microalgae research in the world, Europe, and the European Atlantic area. Marine Drugs 18: 79.
Saldarriaga, J. F., F.J.R. Taylor, P.J. Keeling & T. Cavalier-Smith, 2001. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. Journal of Molecular Evolution 53: 204-213.
Stoecker, D.K., M.D. Johnson, C. de Vargas & F. Not. 2009. Acquired phototrophy in aquatic protists. Aquatic Microbial Ecology 57: 279-310.
Sun, J., J. Ni & Y.S. Ho. 2011. Scientometric analysis of coastal eutrophication research during the period of 1993 to 2008. Environment Development and Sustainability. 13: 353-366.
Takano, Y., G. Hansen, D. Fujita & T. Horiguchi. 2008. Serial replacement of diatom endosymbionts in two freshwater dinoflagellates, Peridiniopsis spp. (Peridiniales, Dinophyceae). Phycologia 47: 41-53.
Takano, Y., H. Yamaguchi, I. Inouye, Ø. Moestrup & T. Horiguchi. 2014. Phylogeny of five species of Nusuttodinium gen. nov. (Dinophyceae), a genus of unarmoured kleptoplastidic dinoflagellates. Protist 165: 759-778.
Yamada, N., J.J. Bolton, R. Trobajo, D.G. Mann, P. Dąbek, A. Witkowski, R. Onuma, T. Horiguchi & P.G. Kroth. 2019. Discovery of a kleptoplastic ‘dinotom’ dinoflagellate and the unique nuclear dynamics of converting kleptoplastids to permanent plastids. Scientific Reports 9:10474.
Yamada, N., S.D. Sym & T. Horiguchi. 2017. Identification of highly divergent diatom-derived chloroplasts in dinoflagellates, including a description of Durinskia kwazulunatalensis sp. nov. (Peridiniales, Dinophyceae). Molecular Biology and Evolution 34:1335-1351.
Yu, J.J., M.H. Wang, M. Xu & Y.S. Ho. 2012. A bibliometric analysis of research papers published on photosynthesis: 1992–2009. Photosynthetica 50: 5-14.
Zhang, Y., J. Tao, J. Wang, L. Ding, C. Ding, Y. Li, Q. Zhou, D. Li & H. Zhang. 2019. Trends in diatom research since 1991 based on topic modeling. Microorganisms 7: 213.
Zhang, Z., B.R. Green & T. Cavalier-Smith. 1999. Single gene circle in dinoflagellate chloroplast genomes. Nature 400:155-159.
Sometido: 25 de octubre de 2021.
Revisado: 24 de noviembre de 2021 (Dos revisores anónimos).
Corregido: 3 de diciembre de 2021.
Aceptado: 6 de diciembre de 2021